之前想写一个函数来计算一个三维物体的最邻近间隙距离并进行可视化。发现Grok提供的代码最简洁,可视化效果最好。这里具体记录下这种99%的工作量依靠AI完成的编程过程。
生成代码用到的prompt详见👆。使用 Grok 3 得到的第一版代码如下,我仅修改了测试数据,并打印执行耗时。
import numpy as np
import pandas as pd
from skimage import io, measure
from scipy.spatial import distance
from scipy.ndimage import distance_transform_edt
import plotly.graph_objects as go
from itertools import combinations
import time
def compute_nearest_distances(mask_path):
# Read the mask image
mask = io.imread(mask_path)
# Get unique labels (excluding background 0)
labels = np.unique(mask)[1:] # Exclude 0 (background)
# Initialize lists for DataFrame
data = {
'label': [], 'xc': [], 'yc': [], 'zc': [],
'd': [], 'label2': [], 'xc2': [], 'yc2': [], 'zc2': [],
'xe1': [], 'ye1': [], 'ze1': [], 'xe2': [], 'ye2': [], 'ze2': []
}
# Compute properties for all regions
props = measure.regionprops(mask)
label_to_prop = {prop.label: prop for prop in props if prop.label > 0}
# Function to compute boundary points
def get_boundary_points(label_mask):
# Create binary mask for the object
binary = (label_mask > 0).astype(np.uint8)
# Compute distance transform
dist = distance_transform_edt(binary)
# Boundary is where distance is small (e.g., <=1)
boundary = (dist <= 1) & binary
return np.where(boundary)
# Process each pair of objects
for label1, label2 in combinations(labels, 2):
# Get binary masks for both objects
mask1 = (mask == label1).astype(np.uint8)
mask2 = (mask == label2).astype(np.uint8)
# Get boundary points
z1, y1, x1 = get_boundary_points(mask1)
coords1 = np.vstack((x1, y1, z1)).T
z2, y2, x2 = get_boundary_points(mask2)
coords2 = np.vstack((x2, y2, z2)).T
# Compute pairwise distances between boundary points
dist_matrix = distance.cdist(coords1, coords2, 'euclidean')
min_dist = dist_matrix.min()
min_idx = dist_matrix.argmin()
idx1, idx2 = np.unravel_index(min_idx, dist_matrix.shape)
# Get closest boundary points
point1 = coords1[idx1]
point2 = coords2[idx2]
# Get centroid for both objects
prop1 = label_to_prop[label1]
prop2 = label_to_prop[label2]
centroid1 = prop1.centroid # (z, y, x)
centroid2 = prop2.centroid
# Store data for label1
data['label'].append(label1)
data['xc'].append(centroid1[2])
data['yc'].append(centroid1[1])
data['zc'].append(centroid1[0])
data['d'].append(min_dist)
data['label2'].append(label2)
data['xc2'].append(centroid2[2])
data['yc2'].append(centroid2[1])
data['zc2'].append(centroid2[0])
data['xe1'].append(point1[0])
data['ye1'].append(point1[1])
data['ze1'].append(point1[2])
data['xe2'].append(point2[0])
data['ye2'].append(point2[1])
data['ze2'].append(point2[2])
# Store data for label2 (symmetric)
data['label'].append(label2)
data['xc'].append(centroid2[2])
data['yc'].append(centroid2[1])
data['zc'].append(centroid2[0])
data['d'].append(min_dist)
data['label2'].append(label1)
data['xc2'].append(centroid1[2])
data['yc2'].append(centroid1[1])
data['zc2'].append(centroid1[0])
data['xe1'].append(point2[0])
data['ye1'].append(point2[1])
data['ze1'].append(point2[2])
data['xe2'].append(point1[0])
data['ye2'].append(point1[1])
data['ze2'].append(point1[2])
# Create DataFrame
df = pd.DataFrame(data)
# For each label, select the row with minimum distance
df = df.loc[df.groupby('label')['d'].idxmin()]
# Reset index
df = df.reset_index(drop=True)
return df, mask, label_to_prop
def visualize_nearest_distances(df, mask, label_to_prop):
fig = go.Figure()
# Colors for different objects
colors = ['red', 'blue', 'green', 'yellow', 'purple', 'orange', 'cyan', 'magenta']
# Plot each object
for idx, label in enumerate(df['label']):
# Get binary mask for the object
binary_mask = (mask == label).astype(np.uint8)
# Get surface points using marching cubes
verts, faces, _, _ = measure.marching_cubes(binary_mask, level=0.5)
# Create surface plot
fig.add_trace(go.Mesh3d(
x=verts[:, 2], y=verts[:, 1], z=verts[:, 0],
i=faces[:, 0], j=faces[:, 1], k=faces[:, 2],
color=colors[idx % len(colors)],
opacity=0.5,
name=f'Object {label}'
))
# Add label text at centroid
prop = label_to_prop[label]
centroid = prop.centroid
fig.add_trace(go.Scatter3d(
x=[centroid[2]], y=[centroid[1]], z=[centroid[0]],
mode='text',
text=[f'Label {label}'],
textposition='middle center',
showlegend=False
))
# Add line for nearest distance
row = df[df['label'] == label].iloc[0]
fig.add_trace(go.Scatter3d(
x=[row['xe1'], row['xe2']],
y=[row['ye1'], row['ye2']],
z=[row['ze1'], row['ze2']],
mode='lines',
line=dict(color='black', width=5),
name=f'Distance {label}-{row["label2"]}'
))
# Update layout
fig.update_layout(
scene=dict(
xaxis_title='X',
yaxis_title='Y',
zaxis_title='Z',
aspectmode='data'
),
title='3D Objects with Nearest Neighbor Distances'
)
return fig
# Example usage
if __name__ == "__main__":
# Path to your mask.tif file
mask_path = 'test-mask.tif'
t0 = time.time()
# Compute distances and get DataFrame
df, mask, label_to_prop = compute_nearest_distances(mask_path)
print(f"Time usage: {time.time()-t0:.2f} s.") # Time usage: 86.95 s.
# Print DataFrame
# print(df)
df.to_csv("test-mask-gap-distance-results.csv", index=None)
# Create and show visualization
fig = visualize_nearest_distances(df, mask, label_to_prop)
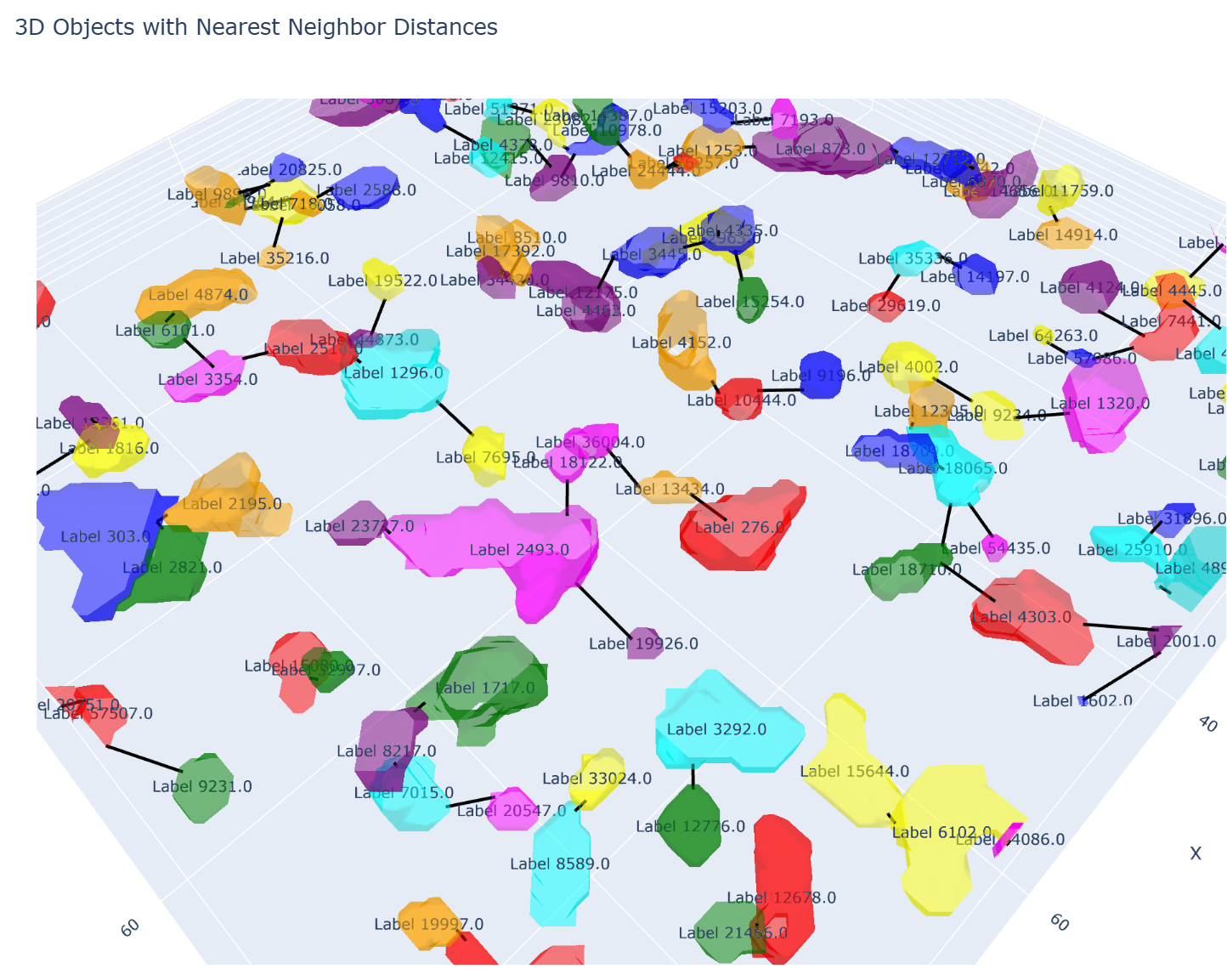
fig.show()这段代码在我的机器(AMD R9 7900X)中执行耗时 86.9秒,这个有点难以接受,因为 test-mask.tif 的shape是(9,100,100),而我真正待分析的数据是 (9, 2560, 2560),所以必须要优化性能的。
然后我就继续提交prompt:
如何优化下面这段代码以提升性能:
...(code)...Grok很快就更新了一般代码,我再复制下来尝试运行,发现报错。但是没关系,我把报错直接复制粘贴反馈给Grok,不需要任何添油加醋。如:
TypeError: regionprops() got an unexpected keyword argument 'properties'
joblib.externals.loky.process_executor.BrokenProcessPool: A task has failed to un-serialize. Please ensure that the arguments of the function are all picklable.
解决上述两个报错后,代码就能跑了。使用测试数据测性能,耗时仅需 3.37 秒。更新后的代码如下:
import numpy as np
import pandas as pd
from skimage import io, measure
from scipy.spatial import KDTree
from scipy.ndimage import binary_erosion
from joblib import Parallel, delayed
from itertools import combinations
import plotly.graph_objects as go
import time
def get_boundary_points(label_mask, max_points=1000):
binary = (label_mask > 0).astype(np.uint8)
eroded = binary_erosion(binary, iterations=1)
boundary = binary & ~eroded
z, y, x = np.where(boundary)
if len(x) > max_points:
indices = np.random.choice(len(x), max_points, replace=False)
x, y, z = x[indices], y[indices], z[indices]
return z, y, x
def process_pair(label1, label2, mask1, mask2, centroid1, centroid2):
# Compute boundary points
z1, y1, x1 = get_boundary_points(mask1)
coords1 = np.vstack((x1, y1, z1)).T
z2, y2, x2 = get_boundary_points(mask2)
coords2 = np.vstack((x2, y2, z2)).T
# Use KDTree for efficient distance calculation
tree = KDTree(coords2)
min_dist, idx2 = tree.query(coords1)
idx1 = np.argmin(min_dist)
min_dist = min_dist[idx1]
point1 = coords1[idx1]
point2 = coords2[idx2[idx1]]
# Return data for both directions
return [
(label1, centroid1[2], centroid1[1], centroid1[0], min_dist, label2, centroid2[2], centroid2[1], centroid2[0], point1[0], point1[1], point1[2], point2[0], point2[1], point2[2]),
(label2, centroid2[2], centroid2[1], centroid2[0], min_dist, label1, centroid1[2], centroid1[1], centroid1[0], point2[0], point2[1], point2[2], point1[0], point1[1], point1[2])
]
def compute_nearest_distances(mask_path):
# Read the mask image
mask = io.imread(mask_path)
labels = np.unique(mask)[1:] # Exclude background
# Compute region properties (compatible with older scikit-image versions)
props = measure.regionprops(mask)
label_to_prop = {prop.label: prop for prop in props if prop.label > 0}
# Precompute binary masks and centroids
label_masks = {label: (mask == label).astype(np.uint8) for label in labels}
label_to_centroid = {label: prop.centroid for label, prop in label_to_prop.items()}
# Parallel processing
results = Parallel(n_jobs=-1, backend='loky')(
delayed(process_pair)(
label1,
label2,
label_masks[label1],
label_masks[label2],
label_to_centroid[label1],
label_to_centroid[label2]
)
for label1, label2 in combinations(labels, 2)
)
# Collect results
n_pairs = len(results) * 2
data = np.zeros((n_pairs, 15), dtype=np.float64)
idx = 0
for result in results:
data[idx:idx+2] = result
idx += 2
# Create DataFrame
df = pd.DataFrame(data, columns=['label', 'xc', 'yc', 'zc', 'd', 'label2', 'xc2', 'yc2', 'zc2', 'xe1', 'ye1', 'ze1', 'xe2', 'ye2', 'ze2'])
df = df.loc[df.groupby('label')['d'].idxmin()].reset_index(drop=True)
return df, mask, label_to_prop
def visualize_nearest_distances(df, mask, label_to_prop):
fig = go.Figure()
# Colors for different objects
colors = ['red', 'blue', 'green', 'yellow', 'purple', 'orange', 'cyan', 'magenta']
# Plot each object
for idx, label in enumerate(df['label']):
# Get binary mask for the object
binary_mask = (mask == label).astype(np.uint8)
# Get surface points using marching cubes
verts, faces, _, _ = measure.marching_cubes(binary_mask, level=0.5)
# Create surface plot
fig.add_trace(go.Mesh3d(
x=verts[:, 2], y=verts[:, 1], z=verts[:, 0],
i=faces[:, 0], j=faces[:, 1], k=faces[:, 2],
color=colors[idx % len(colors)],
opacity=0.5,
name=f'Object {label}'
))
# Add label text at centroid
prop = label_to_prop[label]
centroid = prop.centroid
fig.add_trace(go.Scatter3d(
x=[centroid[2]], y=[centroid[1]], z=[centroid[0]],
mode='text',
text=[f'Label {label}'],
textposition='middle center',
showlegend=False
))
# Add line for nearest distance
row = df[df['label'] == label].iloc[0]
fig.add_trace(go.Scatter3d(
x=[row['xe1'], row['xe2']],
y=[row['ye1'], row['ye2']],
z=[row['ze1'], row['ze2']],
mode='lines',
line=dict(color='black', width=5),
name=f'Distance {label}-{row["label2"]}'
))
# Update layout
fig.update_layout(
scene=dict(
xaxis_title='X',
yaxis_title='Y',
zaxis_title='Z',
aspectmode='data'
),
title='3D Objects with Nearest Neighbor Distances'
)
return fig
# Example usage
if __name__ == "__main__":
# Path to your mask.tif file
mask_path = 'test-mask.tif'
t0 = time.time()
# Compute distances and get DataFrame
df, mask, label_to_prop = compute_nearest_distances(mask_path)
print(f"Time usage: {time.time()-t0:.2f} s.")
# Time usage: 3.4 s for 9x100x100, 322.2 s for 9x256x256
# Print DataFrame
# print(df)
df.to_csv("test-mask-gap-distance-results.csv", index=None)
# Create and show visualization
fig = visualize_nearest_distances(df, mask, label_to_prop)
fig.show()计算结果和可视化效果都和之前的一模一样。
经测试,输入mask.tif包含9x256x256这么多像素,具备一定可用性。如果是9x2560x2560会报内存错误。不过后面可以进一步优化,比如使用移动窗口的方式遍历全局,或者是按对象遍历,应该还是具有较好的可用性的。
我和Grok具体的聊天记录详见此链接(需科学上网)。

此处评论已关闭